A twitter explainer on our latest study, Inherited DNA Repair Defects in Colorectal Cancer – congrats @s_aldubayan @MattYurgelun @SapnaSyngal et al! @DanaFarber @broadinstitute @DamonRunyon @AJHGNews authors.elsevier.com/a/1Web9geWqOxr 
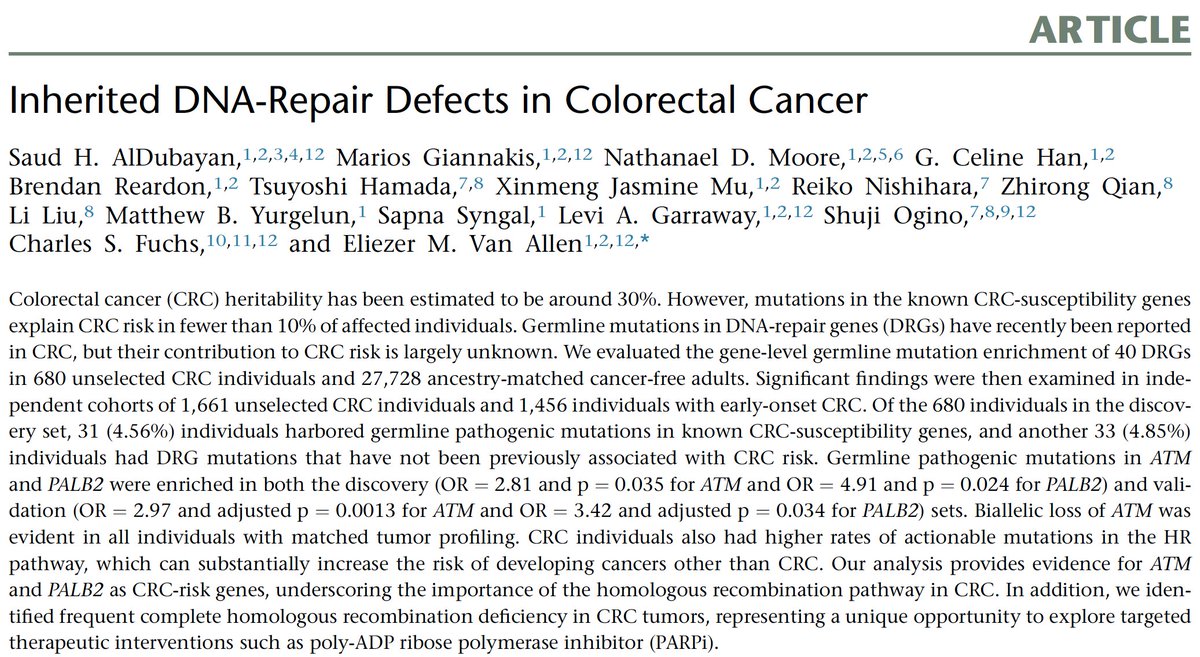
There’s an established heritability for colorectal cancer hovering around 30%, though only roughly 5-10% is explained by known syndromes involving genes like APC, MSH2, and PTEN, among others – the rest is so-called “missing heritability” 
In addition, traditional clinical measures like family history have not thus far been closely related to the identification of bona fide pathogenic mutations, so new studies are needed looking at a broader patient population to understand the patterns of these mutations
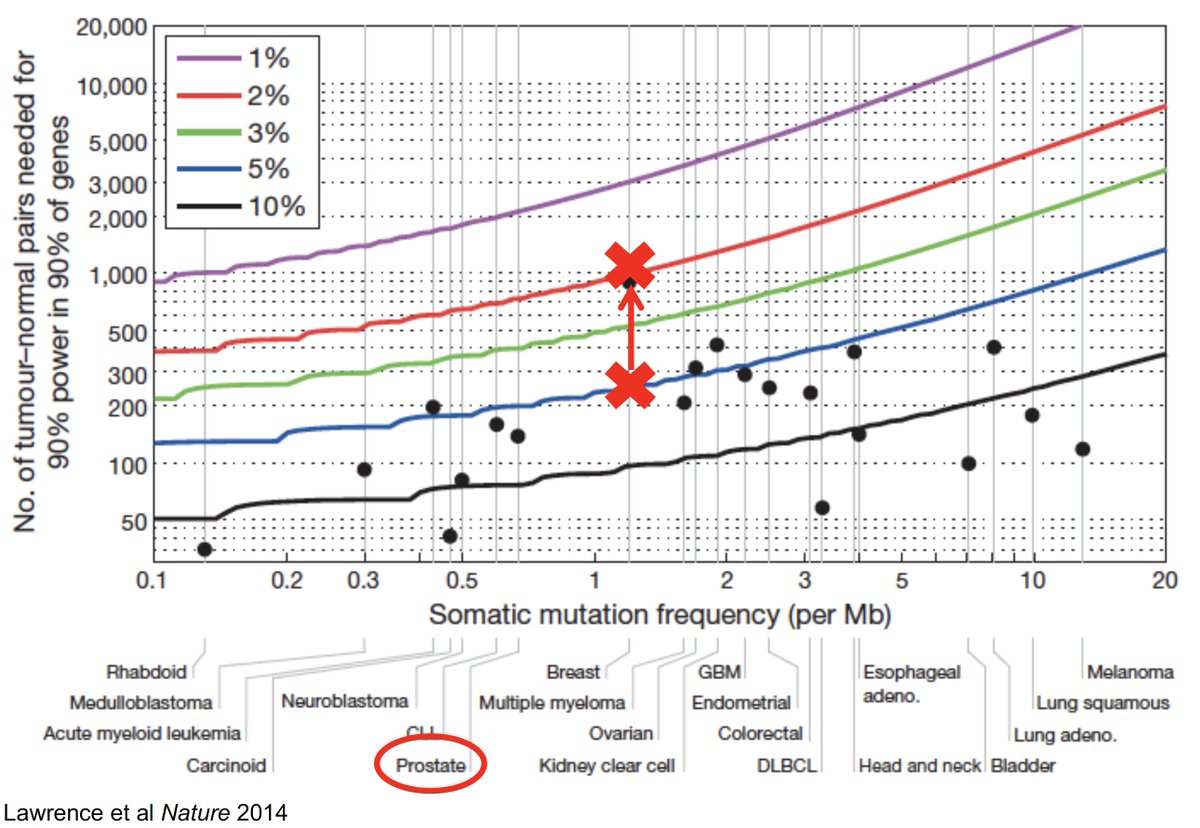
We were lucky to work with Pritchard, @quimmateo et al to establish an approach to looking at DNA repair germline mutations in #prostatecancer (nejm.org/doi/full/10.10…), so we wondered whether similar patterns were present in colon cancer using a case/control approach
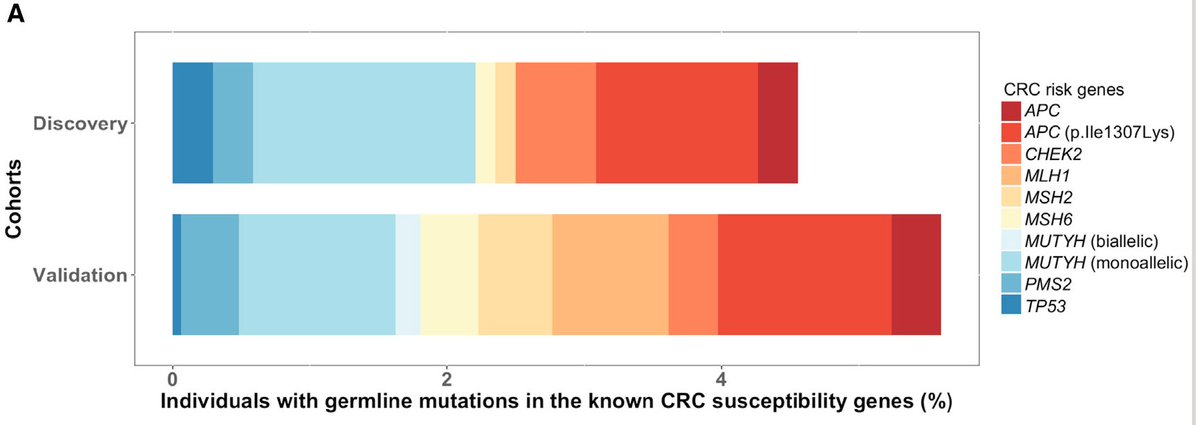
In a cohort of 680 germline exomes from colon cancer patients, there was enrichment of bona fide pathogenic mutations in ATM and PALB2 when compared to a large cohort of ancestry-matched cancer-free adults 
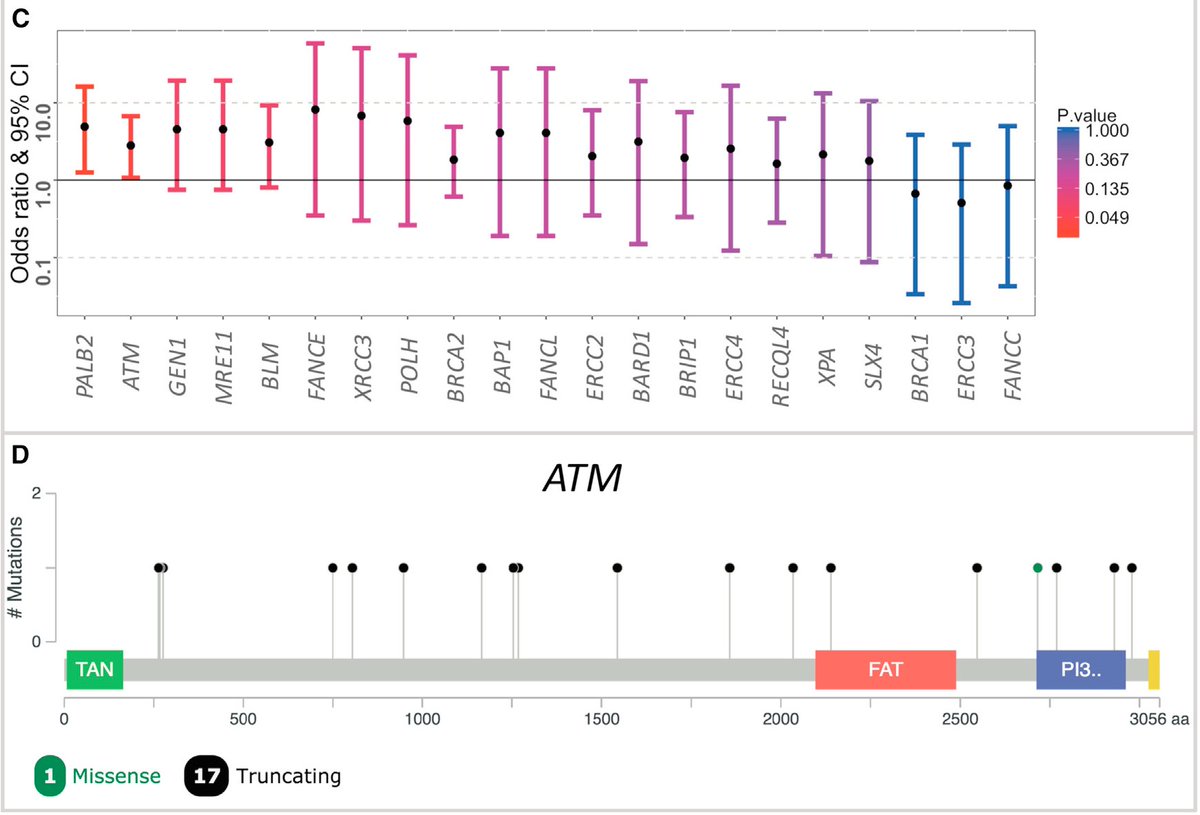
Tangentially, here is example number 2903840932 whereby @exac_exomes can have such a profound impact in areas of research that may have been hard to predict at the outset – thx again @dgmacarthur et al
We then extended this observation into thousands of unselected colon cancer cases (either typical or early onset cases), and where available looked at the somatic status of these genes in these patients 

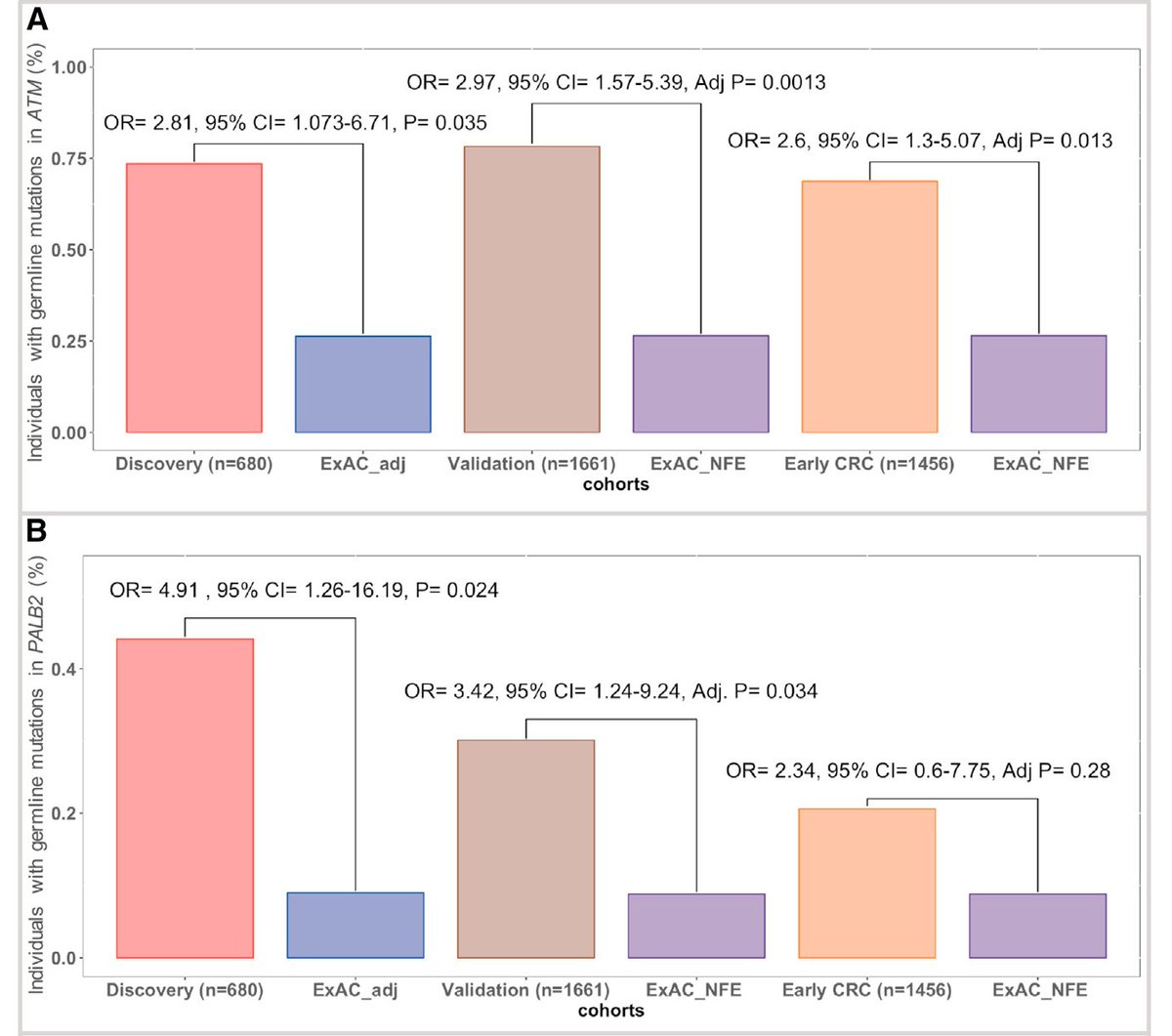
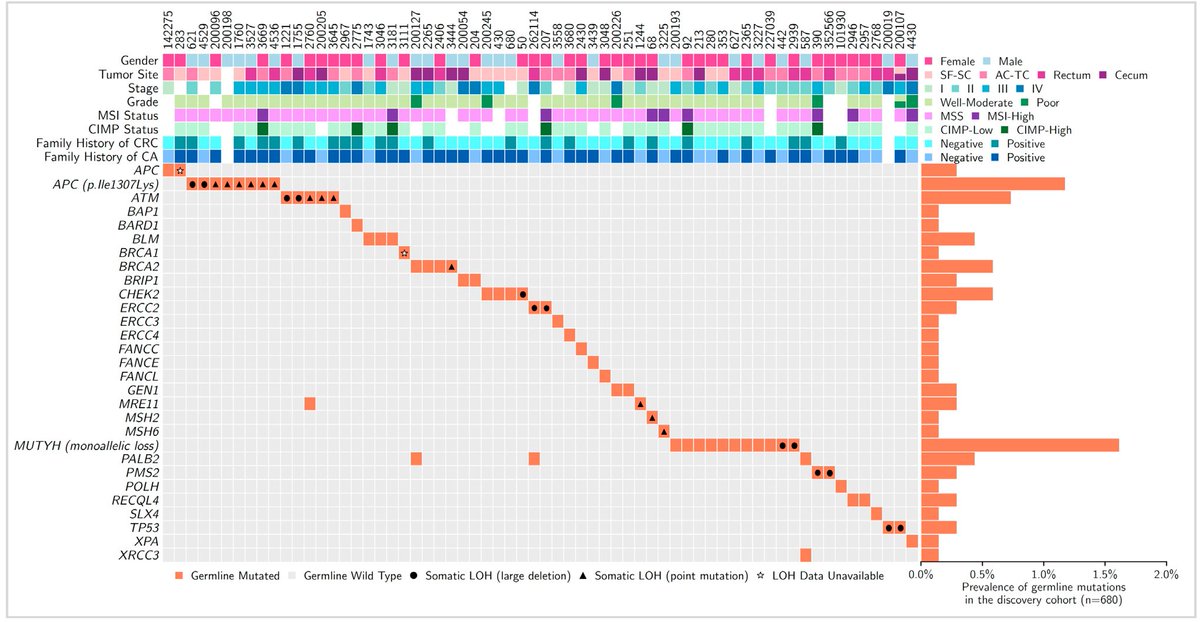
Overall, this study builds on prior work to (we think) formally establish ATM and PALB2 as colon cancer risk genes - see also:
ascopubs.org/doi/abs/10.120…
jamanetwork.com/journals/jamao…
ascopubs.org/doi/abs/10.120…
jamanetwork.com/journals/jamao…
As a result, these findings may have potentially immediate practice changing implications re: genetic testing, and may open pathways for usage of therapies like PARP inhibitors in genetically defined colon cancer patient populations
Lastly, it’s worth noting we did not generate a single new exome for this study, but rather asked questions using data generated for other purposes - this has helped teach us on effective strategies for doing this prospectively to ask other clinically relevant questions <fin>
• • •
Missing some Tweet in this thread? You can try to
force a refresh