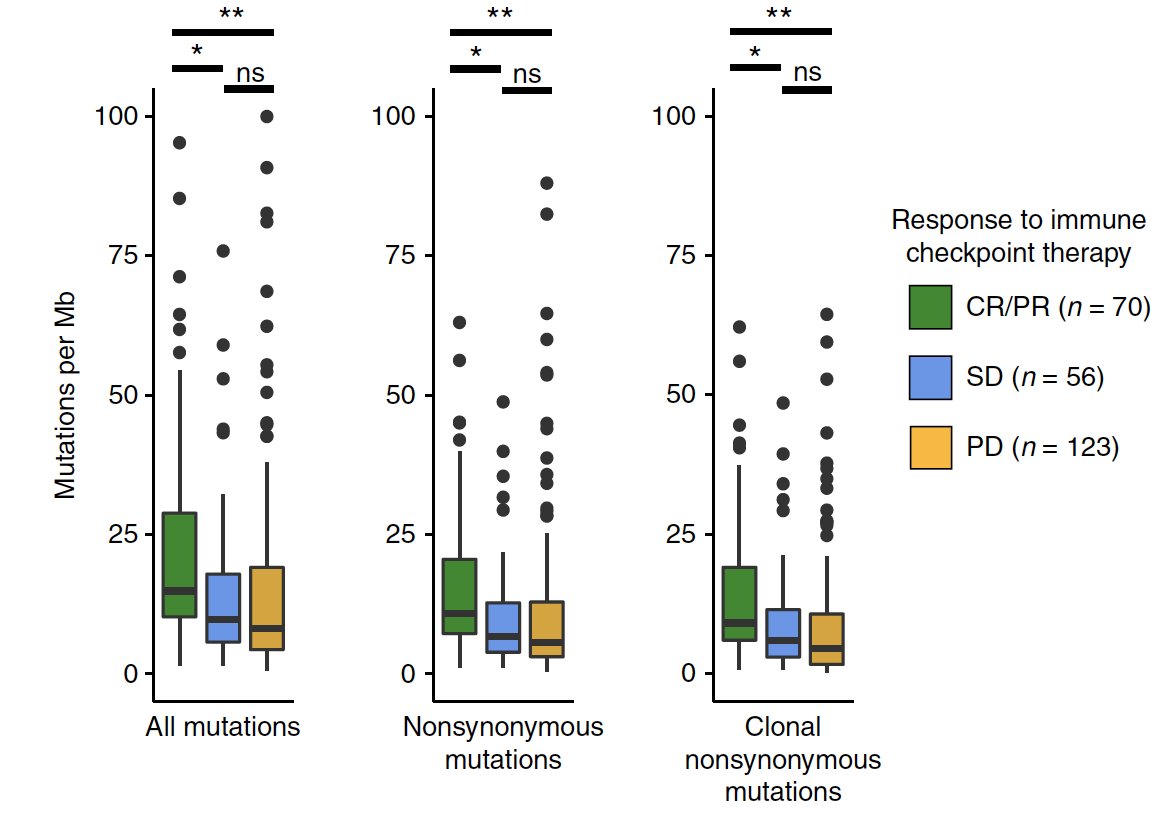
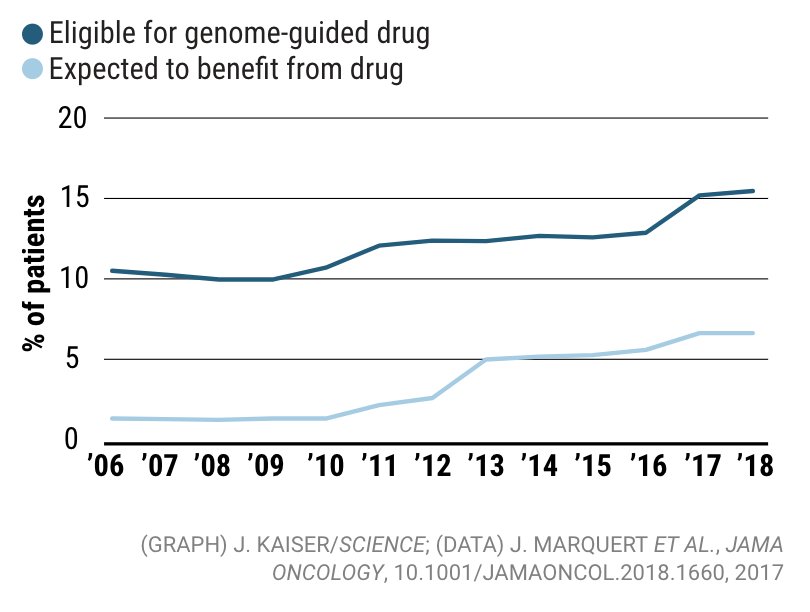
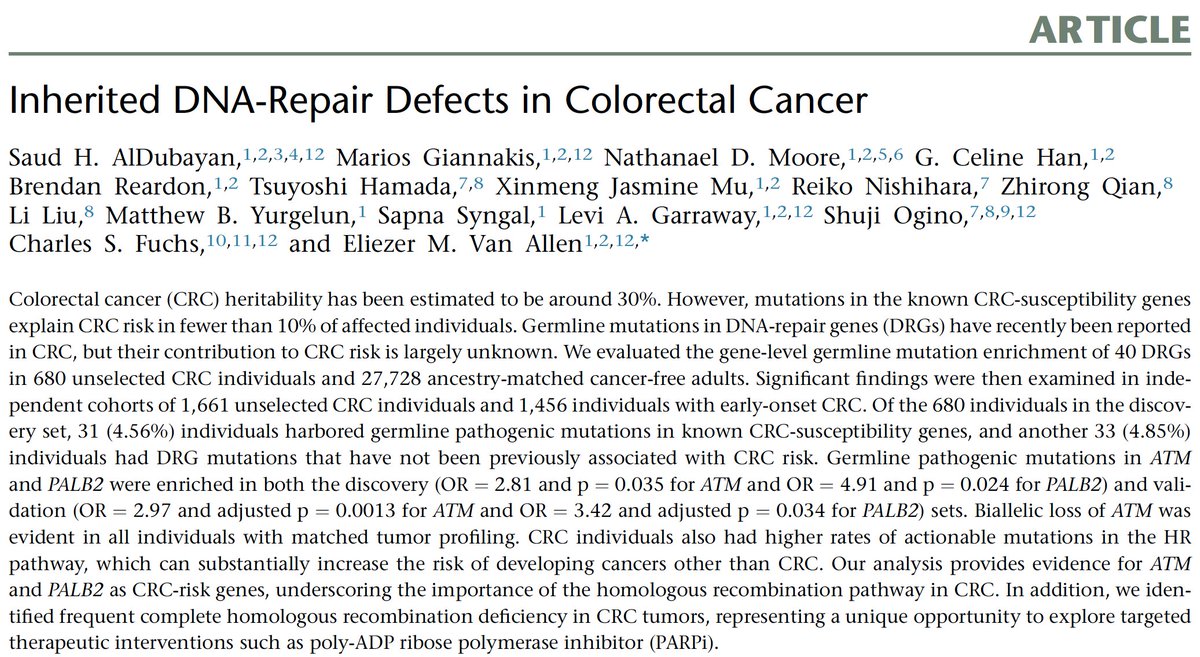
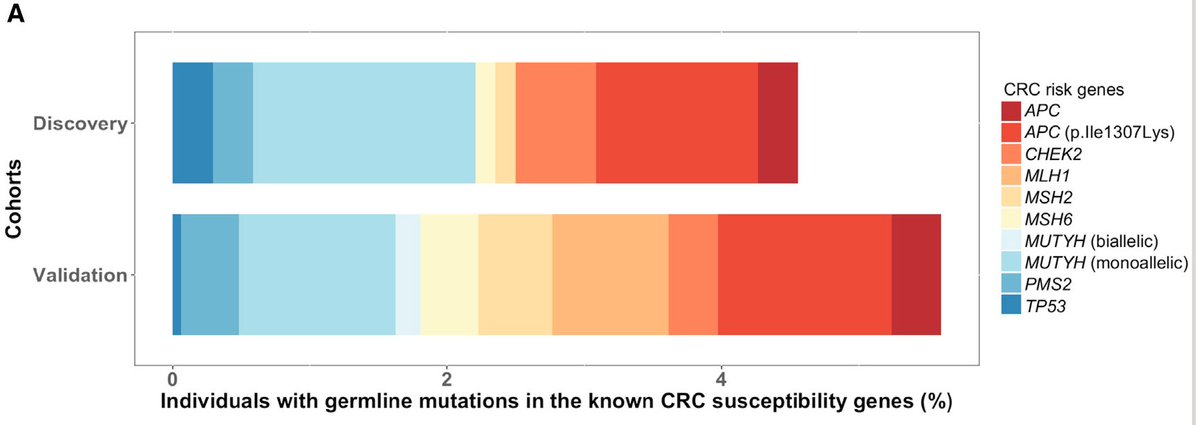
A twitter explainer for our latest #teamscience study rdcu.be/KwgV congrats Josh @stephanie_mul @dliu_ccb @nikolausschultz Arul @CharlesSawyers et al @PCFnews @SU2C @DanaFarber @broadinstitute @MSKCC_OncoNotes @NatureGenet [1/n] 

I've been lucky to grow up academically around many #prostatecancer scientific 'big machers' & helped with early cancer genomic maps over the last 5+ years, e.g.:
1) rdcu.be/Kw8d (@Chris_Barbieri1)
2) cell.com/cell/fulltext/…
3) cell.com/abstract/S0092… (@NCIgenomics)
1) rdcu.be/Kw8d (@Chris_Barbieri1)
2) cell.com/cell/fulltext/…
3) cell.com/abstract/S0092… (@NCIgenomics)
About two years ago, @stephanie_mul tallied the whole exome data we had & mapped it against theoretical power calculations @gaddyg set forth for discovering significant cancer genes in any cancer type (see: ncbi.nlm.nih.gov/pubmed/24390350) 
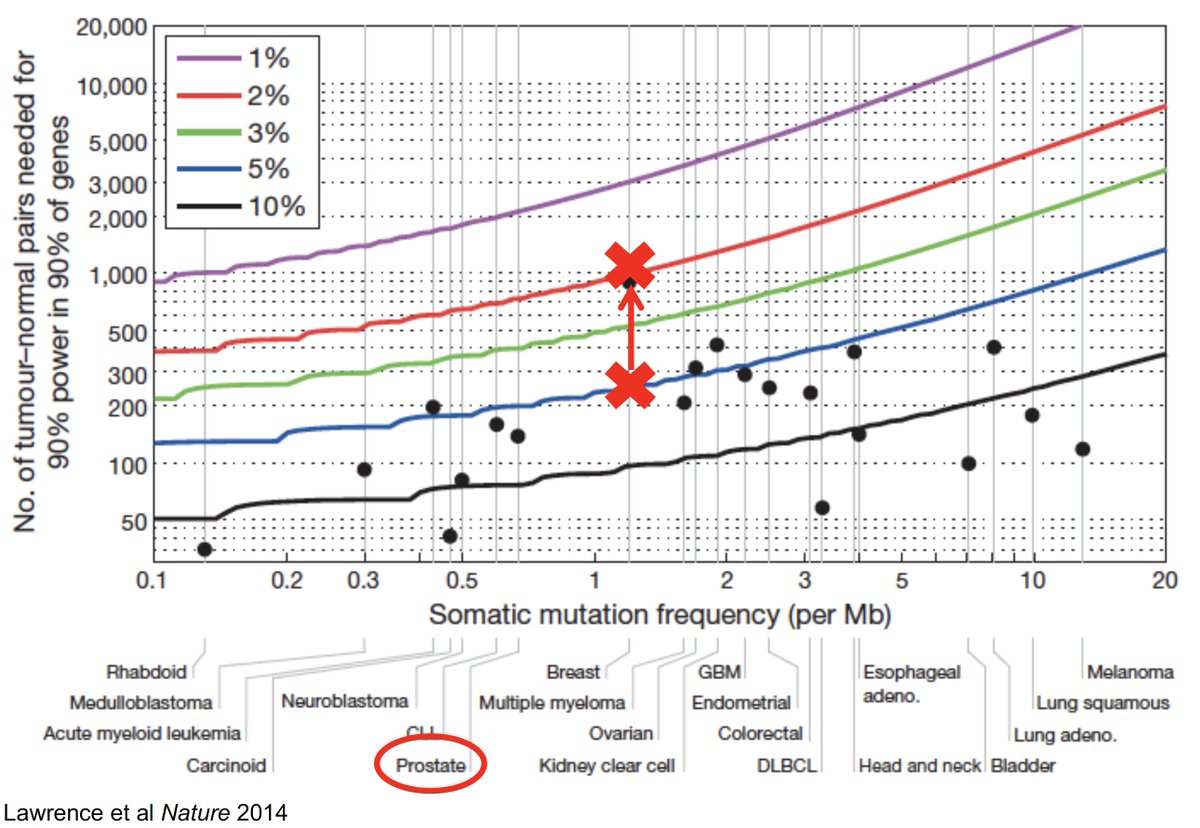
Turned out we had a lot! So we started wondering, "If you have enough tumor genomics in any one cancer type, can you find mutations in genes that invoke *all* Cancer Hallmarks, even those never seen in that cancer?" (see: cell.com/abstract/S0092…) 
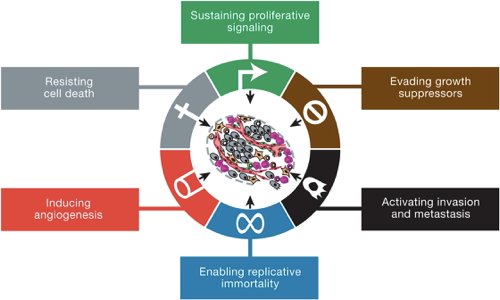
We took a first stab at this at our 2015 PCF/SU2C team meeting → @nikolausschultz approached me at snack table (The Great Chips & Dip Scientific Summit of 2015) → he was doing the same thing, and we both immediately agreed we should do this together because #squadgoals
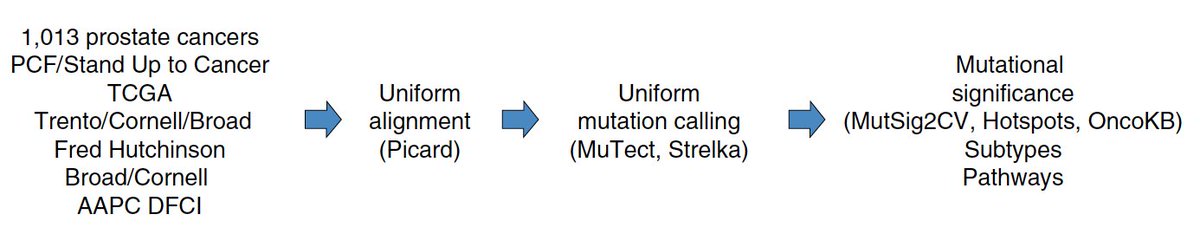
After numerous analyses, realigning, re-re-aligning, re-analysis, etc, we had a refined set of 1,013 #prostatecancer exomes - in 2018, we also now have many cancer genomics & biology priors to apply with a statistical framework (h/t @MarkARubin1) and learn! 
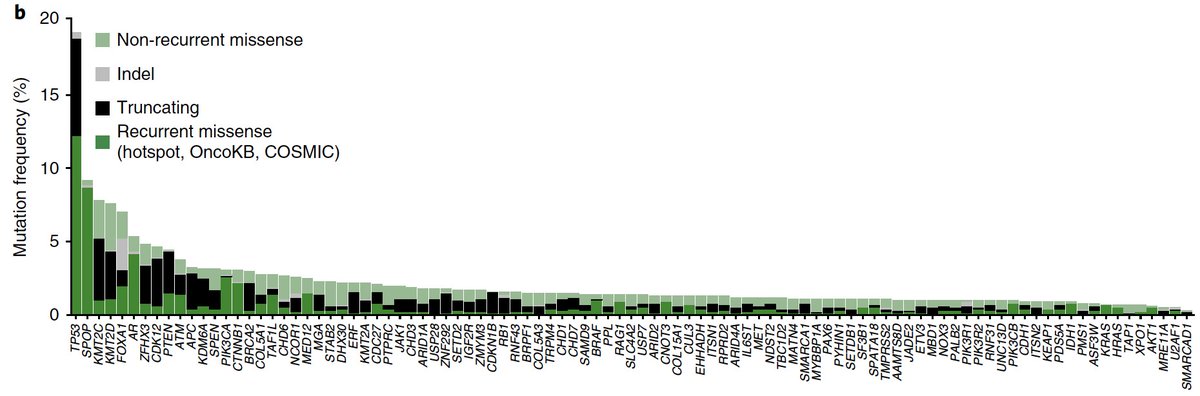
Main finding: A true “long tail” of significantly mutated genes in #prostatecancer, including genes/pathways we had no idea were involved & rare, but helped us cover the remaining territory of the Cancer Hallmarks (e.g. SF3B1/splicing, Ras signaling) 


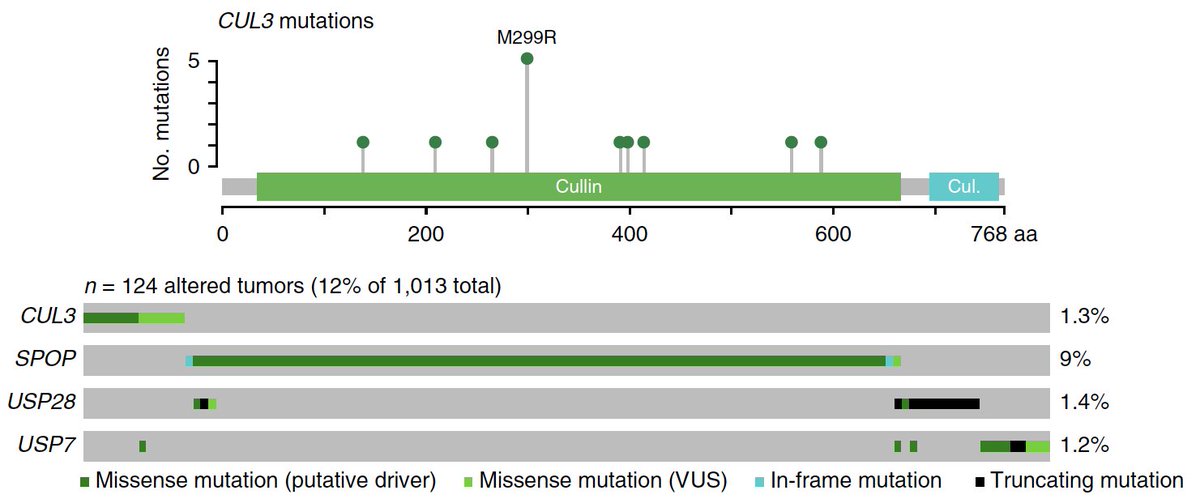
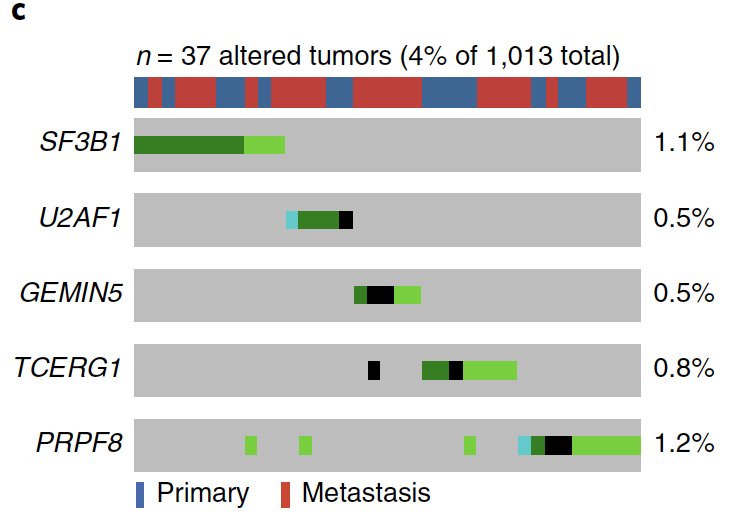
We also saw an entire subclass of prostate cancer with mutations exclusively in epigenetic genes, raising new therapeutic & tumor-immune hypotheses, plus new collabs with @kadochlab @CKadoch et al cc: @SKCCDirector 
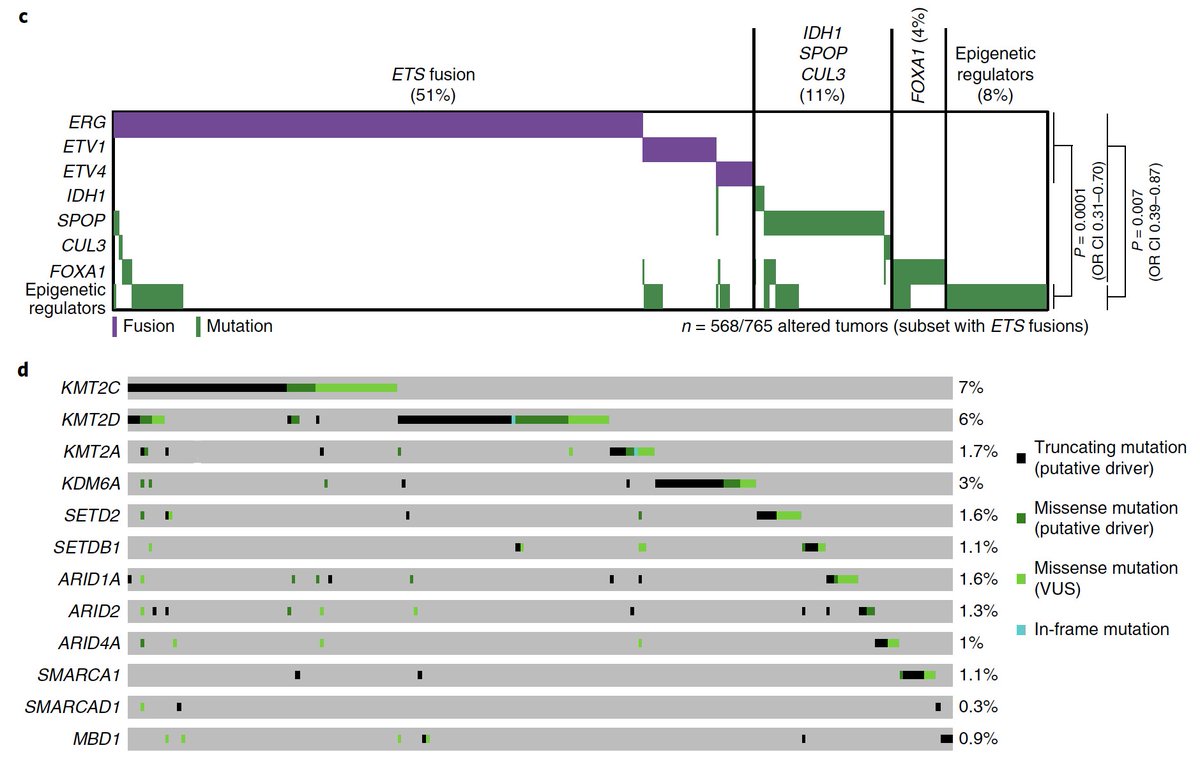
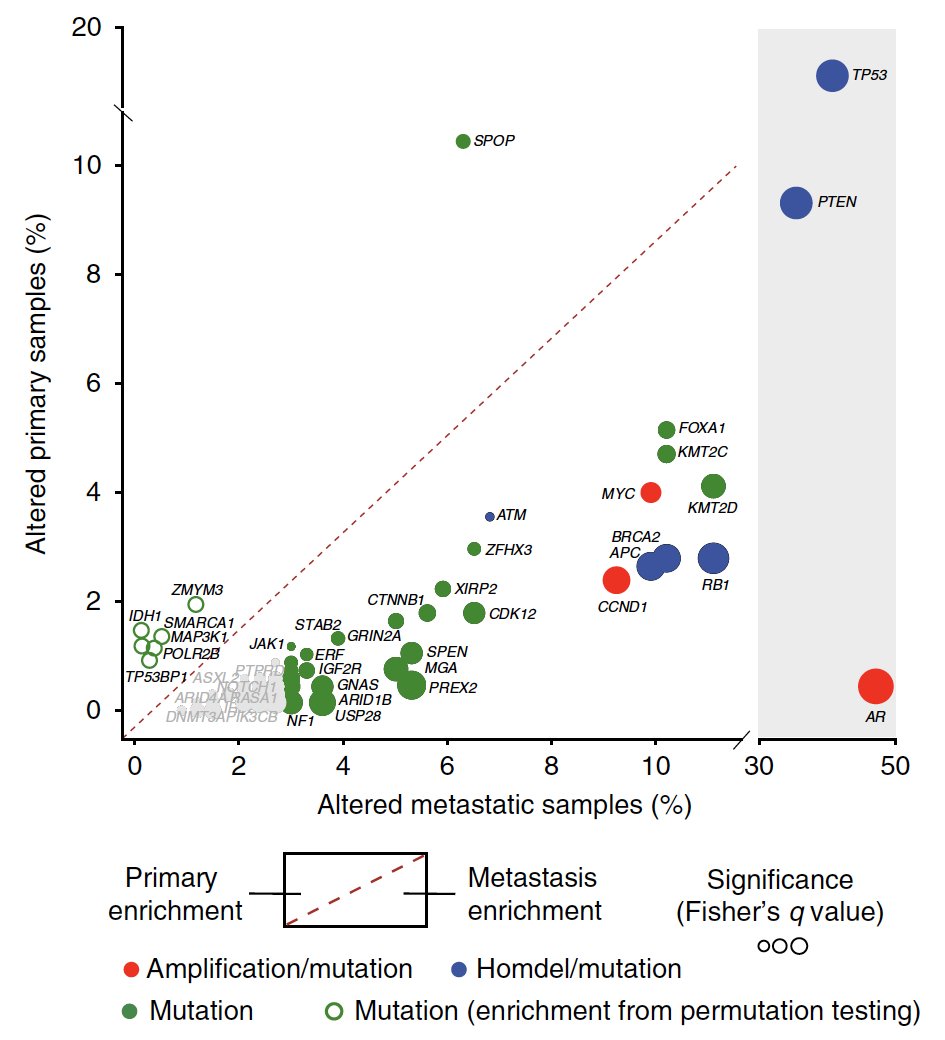
With a bigger cohort, we also could take a better swing at defining a genomic “signature” of metastatic vs. primary prostate cancer – we are still a long way from a clinical test, but the potential is vast, e.g. wsj.com/articles/prost… @LucetteLagnado 
Of course, we hope to augment this cohort (exclusively from academic sites via traditional means) with the patient-driven Metastatic Prostate Cancer Project cohort → 3 months in & 100s of men have said “Count Me In!” mpcproject.org @PrCaProject @corrie_painter et al 
Finally, this is yet another example of the power of #teamscience → just the latest product from a group effort that has yielded numerous findings for #prostatecancer, and we're excited/hopeful for where this will go in the future! @PCFnews @PCF_Science @SU2C [fin]
• • •
Missing some Tweet in this thread? You can try to
force a refresh