Finally the comments to Mitalipov’s group paper on #CRISPR in human embryos are out in @nature as well as the rebuttal from Ma et al. as brief communications arising #paywall. Let’s have a look more in details -> Thread
One of the major finding of the Ma et al. paper was the discovery of high frequency of inter-homologous repair mechanisms aka loss of heterozygocity LOH -> 1 allele is used as a template for
#CRISPR-HDR -> WT alleles are in fact alleles from the maternal alleles only.
#CRISPR-HDR -> WT alleles are in fact alleles from the maternal alleles only.
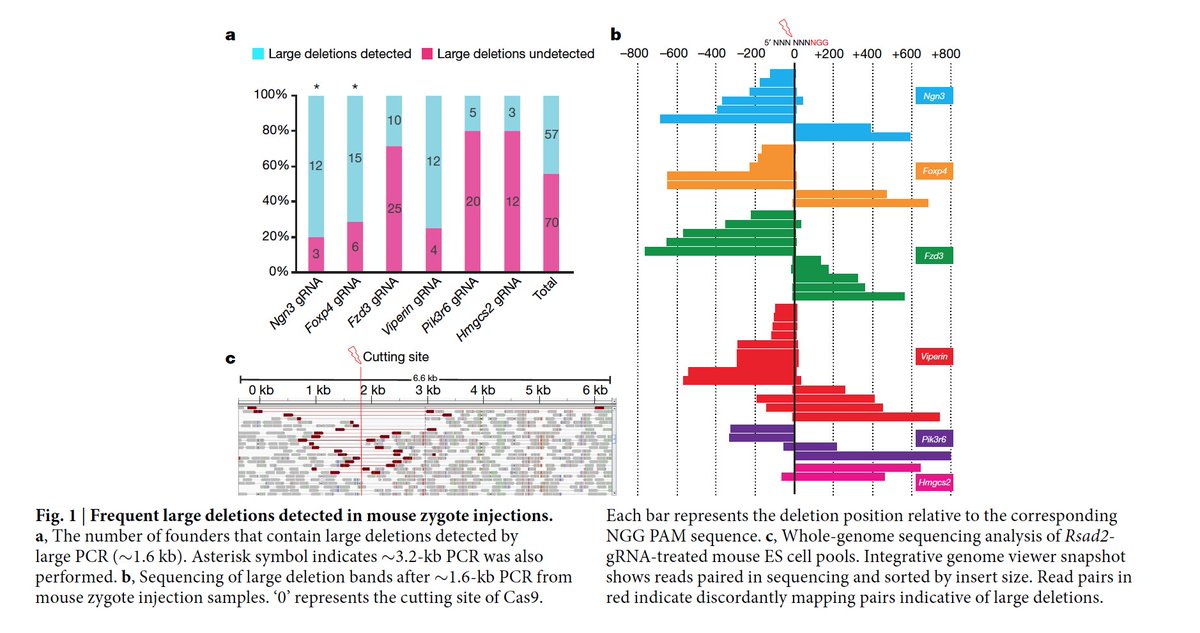
We have been debating a lot on this in the last year or so. The first important comment from @Fatwa_Adikusuma et al is Ma et al. missed deleted alleles. In the paper they edited 6 loci in zygotes and EScells and checked for large deletions nature.com/articles/s4158…
The results are very clear and show high frequency of deletions > 30% of large deletions which is not captured by classical PCRs. This independently confirms large deletions/inversion... from Allan Bradley’s group published recently in @NatureBiotech 
The second comment from Maria Jasin's group raised the fact that inter-homologous recombination occurs late in the development aka 2 cell stage. nature.com/articles/s4158… This was demonstrated in a recent paper in Science science.sciencemag.org/content/361/63…
Therefore The excess of WT embryos observed after CRISPR HDR from Ma et al. were 1) large deletions missed and/or 2) parthenogenesis. As I mentioned before, the evidence from Maria Jasin group and collaborators is really compelling & independently confirmed 

So the rebuttal from Ma et al is fascinating nature.com/articles/s4158…. Regarding the large deletions, Ma et al. provided a series of gels (Fig 1) from long range PCRs from 400 bp to > 10kb only on 8 samples and there is a lot of extra bands! 



So they cut some bands (how many?) and did some Sanger and found nothing specific. In short they found no mosaicism. Ma et al. argued that Cas9 RNP doesn’t generated any mosaicism ≠ mRNA unlike Paul Thomas group paper which is true and I would be partly agreed with this
They haven’t done any sequencing those samples, well only Sanger but not short or long read sequencing. It would have been a compelling argument to formally exclude any mosaicism by Amplicon sequencing TBH. I found astonishing they could get away with it for a @nature paper
@nature The second point is interesting. They look at their WGS data and look for informative SNPs (aka polymorphic SNP between paternal and maternal alleles) and they found 3 SNPs. 

By genotyping the blastomers at ≠ stages they found evidence of inter-homologous repair. However, some of the peaks are really small making me thinking they missed out on detecting mosaic alleles, rather than detecting real inter homologous repair 



Besides mosaicism, I can‘t get my head around these haplotypes & explain this discrepancy between the cell biology observations from Maria Jasin’s group and the haplotypes presented in Ma et al. paper. 



Inter homologous repair might be real, but I really struggle to be convinced from the data presented in Ma et al. rebuttal. I found those results from Ma et al. not convincing at all. Very surprised they could get away with it
Some more general comments now. 1/ I found frustrating having to comment on papers under #paywall and important part of the public debate. These should have been accessible to everyone and be open access
2/ It is a complex debate on the effect of #CRISPR-Cas9 on DSB and I anticipate some headlines on #genetherapy in human embryos using CRISPR, which is not what these 3 papers were about.
• • •
Missing some Tweet in this thread? You can try to
force a refresh